F₁Fₒ Rotation Mechanism
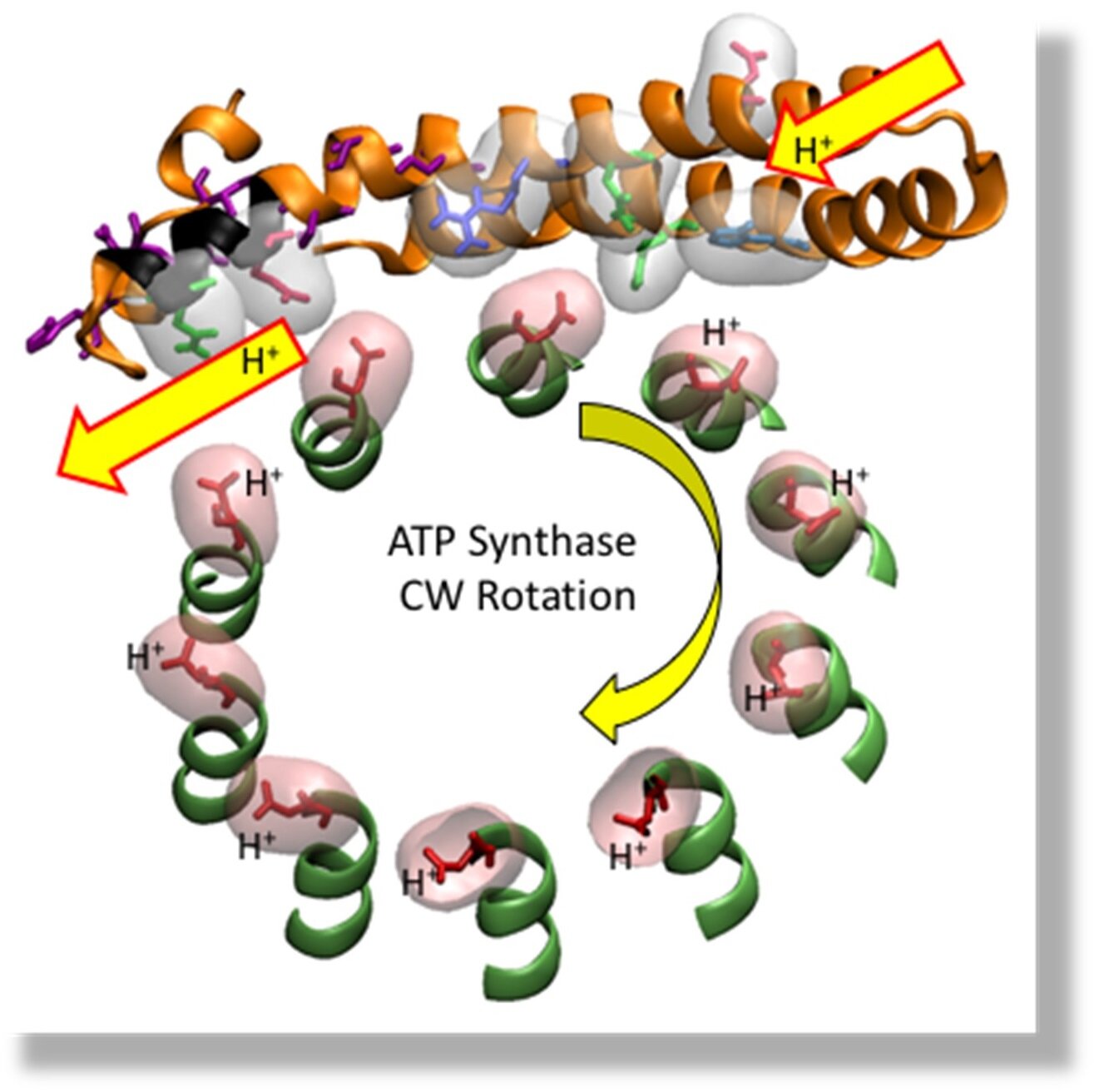
Mechanism of the Fₒ Molecular Motor
Cross-section of E. coli Fₒ showing the c₁₀-ring, and the interface with subunit-a helices that have the input and output proton channels.
In F₁Fₒ, the Fₒ motor uses a transmembrane proton gradient as an energy source to drive rotation in the ATP synthesis (CW) direction against the CCW force generated by the F₁-ATPase.
We resolved the rotational stepping of single c-subunits of the E. coli Fₒ c₁₀-ring and observed that Fₒ subunit-a can push the c-ring in the ATP synthesis direction against the force of F₁ ATPase-dependent rotation.
F₁Fₒ in a lipid bilayer nanodisc (LBN) attached to the slide by His-tags (HT), and to the AuNR at the c-ring via streptavidin (SA).
Single-molecule studies of this motor were limited due to partial dissociation of the subunit-a stator from the c-ring rotor in detergent solubilized preparations. We overcame this problem by incorporating purified F₁Fₒ into lipid nanodiscs that have the physical properties of a biological membrane but are small enough for single molecule studies. The AuNR assay is also required to resolve the rotational stepping of Fₒ.
Counterclockwise rotation during an F₁-ATPase power stroke from an F₁Fₒ molecule where transient dwells (TDs) occur ~36° apart. The first and second TDs for clockwise rotation rotated 10° and 25° (in the ATP synthesis direction).
The single c-subunit steps are evident as transient dwells temporarily stop an F₁-ATPase power stroke for ~125 μs an average of every 36° for the c₁₀-ring. Their occurrence increases with decreasing pH from pH 7 to pH 5, with a distribution that fits to 3 Gaussians that equate to low, medium, and high efficiencies of transient dwell formation. The pH dependence is consistent with differences in the pKa values of the ATP synthase proton input and output channels involved with generating torque on the c-ring.
Distributions of the % of power strokes containing transient dwells (per 300 power stroke data set) at pH 5.0 vs 7.0, and the misalignment of the Fₒ c₁₀-ring with the 120° positions of the F₁ catalytic dwells.
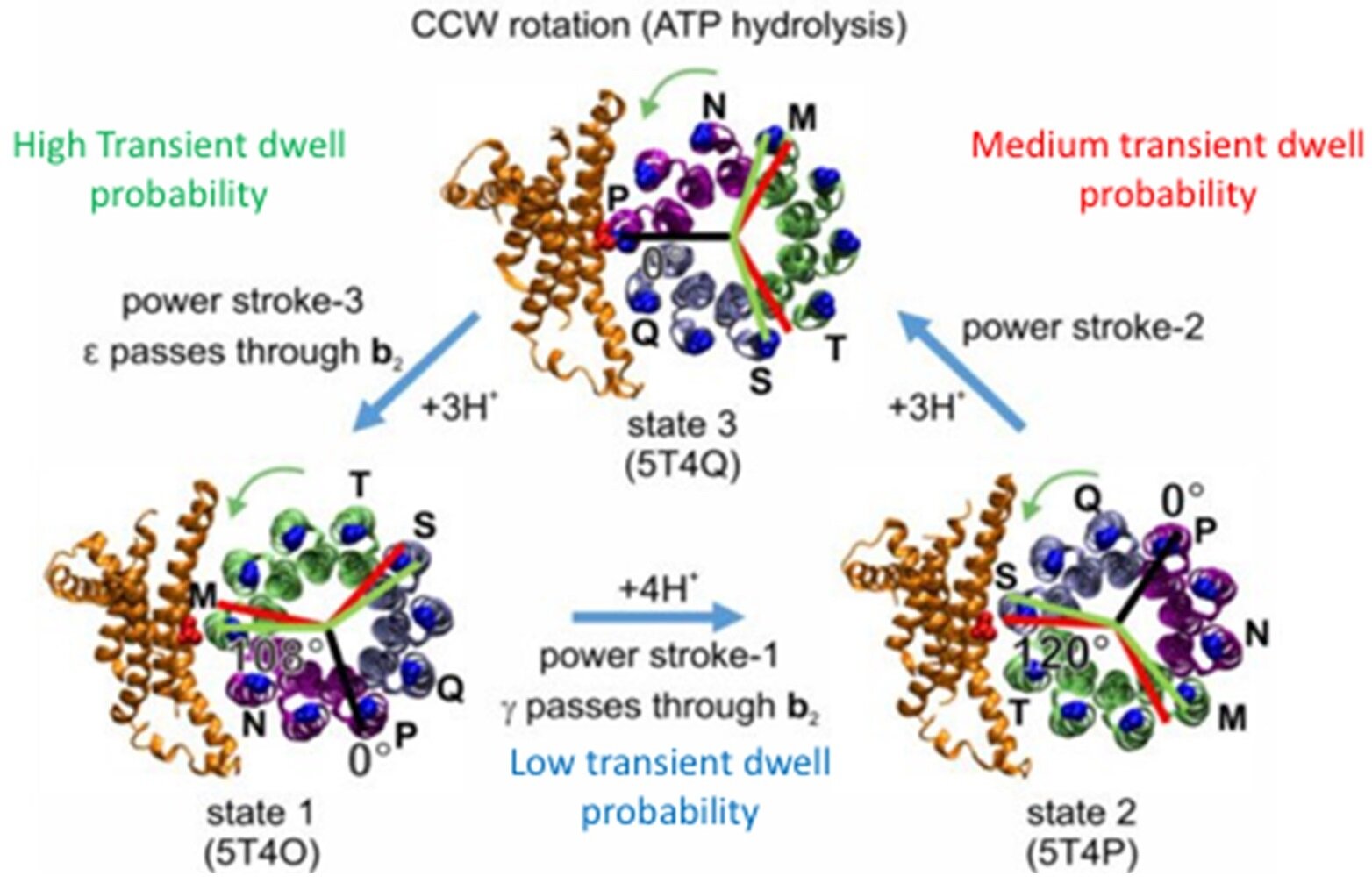
Cryo-EM structures of the three 120° positions of the rotor relative to the stator.
Because F₁ has catalytic dwells 120° apart, and the c₁₀-ring transient dwells occur in 36° steps, the F₁ and Fₒ dwells will differ from the resting c-ring position by +12° or by -12°, which will result in elastic energy that either increases or decreases the ability of the c-ring to rotate CW against the force of the F₁-ATPase generated torque.
Assignment of 120° cryo-EM structures with the high, medium, and low transient dwell efficiencies.
Because we collect and analyze the data from only one of the three power strokes for each single F₁Fₒ molecule, the data from each molecule will represent the asymmetric position of the central rotor with the peripheral stalk where the c-ring is aligned with F₁ or is either in the +12° or the -12° misalignment position. This enabled us to align the low, medium, and high efficiency transient dwell formation with the cryo-EM structures of the three rotary positions of the rotor with the peripheral stalk.
We recently completed a single-molecule study of the effects of mutations of residues involved with proton translocation in the subunit-a channels that has been posted on bioRxiv by the Journal where it is under review. These results are providing important new insight into the molecular mechanism of the Fₒ molecular motor.
Publications
Yanagisawa, S. and Frasch, W.D. (2021) pH-dependent 11° F₁Fₒ ATP synthase sub-steps reveal insight into the Fₒ torque generating mechanism, in review, available on BioRxiv.
Sielaff, H., Yanagisawa, S., Frasch, W.D., Junge, W., and Börsch, M. (2019) Structural Asymmetry and Kinetic Limping of Single Rotary F-ATP Synthases. Molecules 24, 504-529.
Yanagisawa, S. and Frasch, W.D. (2017) Protonation-Dependent Rotation of the F-type ATP synthase c-ring observed by Single-Molecule Measurements, J. Biol. Chem., 292, 17093-17100.
Martin, J., Hudson, J., Hornung, T., and Frasch, W.D. (2015) Fₒ-driven Power Stroke Rotation Occurs against the Force of F₁ATPase-dependent rotation in the FₒF₁ ATP synthase, J. Biol. Chem., 290, 10717-10728.
Spetzler, D., Ishmukhametov, R., Hornung, T., Martin, J., York, J., Jin-Day, L., and Frasch, W. D. (2012) “Energy Transduction by the Two Molecular Motors of the F₁Fₒ ATP Synthase”, in Photosynthesis: Plastid Biology, Energy Conversion and Carbon Assimilation, Eaton-Rye, J. J., Tripathy, B. C., and Sharkey. T. D. eds., Springer, Advances in Photosynthesis and Respiration 34, Dordrecht, The Netherlands, Chapter 22, pp. 561-590.
Ishmukhametov, R., Hornung, T., Spetzler, D., and Frasch, W. D. (2010), Direct Observation of stepped proteolipid ring rotation in E. coli FₒF₁-ATP synthase. EMBO J 29:3911-3923.